- Institute of Hydrobiology, Technische Universität Dresden, Dresden, Germany
The use of antibiotic agents as growth promoters was banned in animal husbandry to prevent the selection and spread of antibiotic resistance. However, in addition to antibiotic agents, heavy metals used in animal farming and aquaculture might promote the spread of antibiotic resistance via co-selection. To investigate which heavy metals are likely to co-select for antibiotic resistance in soil and water, the available data on heavy metal pollution, heavy metal toxicity, heavy metal tolerance, and co-selection mechanisms was reviewed. Additionally, the risk of metal driven co-selection of antibiotic resistance in the environment was assessed based on heavy metal concentrations that potentially induce this co-selection process. Analyses of the data indicate that agricultural and aquacultural practices represent major sources of soil and water contamination with moderately to highly toxic metals such as mercury (Hg), cadmium (Cd), copper (Cu), and zinc (Zn). If those metals reach the environment and accumulate to critical concentrations they can trigger co-selection of antibiotic resistance. Furthermore, co-selection mechanisms for these heavy metals and clinically as well as veterinary relevant antibiotics have been described. Therefore, studies investigating co-selection in environments impacted by agriculture and aquaculture should focus on Hg, Cd, Cu, and Zn as selecting heavy metals. Nevertheless, the respective environmental background has to be taken into account.
Introduction
The number of infections caused by antibiotic resistant bacteria is rising worldwide (Levy and Marshall, 2004). Because of this critical development associated with the loss of the therapeutic potential of antibiotics, some reports are already describing the start of the post-antibiotic era with the consequence of rising difficulties in the treatment of infectious diseases (Alanis, 2005). The decrease of antibiotic use does not necessarily prevent the spread and maintenance of antibiotic resistance in clinical but also in natural environments (Salyers and Amábile-Cuevas, 1997). Therefore, we need to find additional ways to slow down this alarming development (Aminov, 2010). For this reason, it is essential to fully understand the mechanisms and especially the triggers of the evolution and dissemination of antibiotic resistance.
Since there is evidence for recent exchanges of antibiotic resistance genes between environmental and pathogenic bacteria (Poirel et al., 2002, 2005; Forsberg et al., 2012), research brought the natural environment into focus. It is likely that the spread and evolution of antibiotic resistance is triggered or catalysed by anthropogenic pollutants. It has been proved that antimicrobial agents different from antibiotics have the ability to promote a co-selection process, indirectly selecting for antibiotic resistance (Baker-Austin et al., 2006). Heavy metal contaminations are widely spread, whereby agriculture (Han et al., 2001) as well as aquaculture (Burridge et al., 2010) contributing to that environmental burden.
Additionally, due to agricultural and aquacultural practice antibiotics are transferred to soil and water environments, for example soil being fertilized with antibiotic containing manure and sewage sludge (Heuer et al., 2011). Once the antibiotics reach soil environments they may leach to water environments (Boxall et al., 2002). Although the use of antibiotics added to fish feed in aquacultures decreased significantly after the development of vaccinations (Sørum, 2006), the medication for fish with antibiotics as feed amendments is still in practice. The discharge of heavy metals together with antibiotics from agriculture and animal production-linked ecosystems to the environment may cause a combined effect of selection and co-selection toward antibiotic resistant bacteria. Therefore, soil and water bodies impacted by agriculture and aquaculture are hot spots of the evolution of antibiotic resistant bacteria and require special scientific consideration.
Here, we review the risk for metal driven co-selection of antibiotic resistance by addressing heavy metal sources as well as heavy metal toxicity, with regard to aquaculture and agriculture. Additionally, we will review co-selection mechanisms and identify heavy metal concentrations that potentially induce antibiotic resistance co-selection.
Farming and Aquaculture as Sources of Heavy Metals
The anthropogenic contamination of the environment with heavy metals is a serious problem. Aquaculture (Burridge et al., 2010) and agricultural practices (Han et al., 2002; Nicholson et al., 2003) contribute to this world wide pollution due to diverse applications of metals in feed additives, organic and inorganic fertilizers, pesticides, and anti-fouling products.
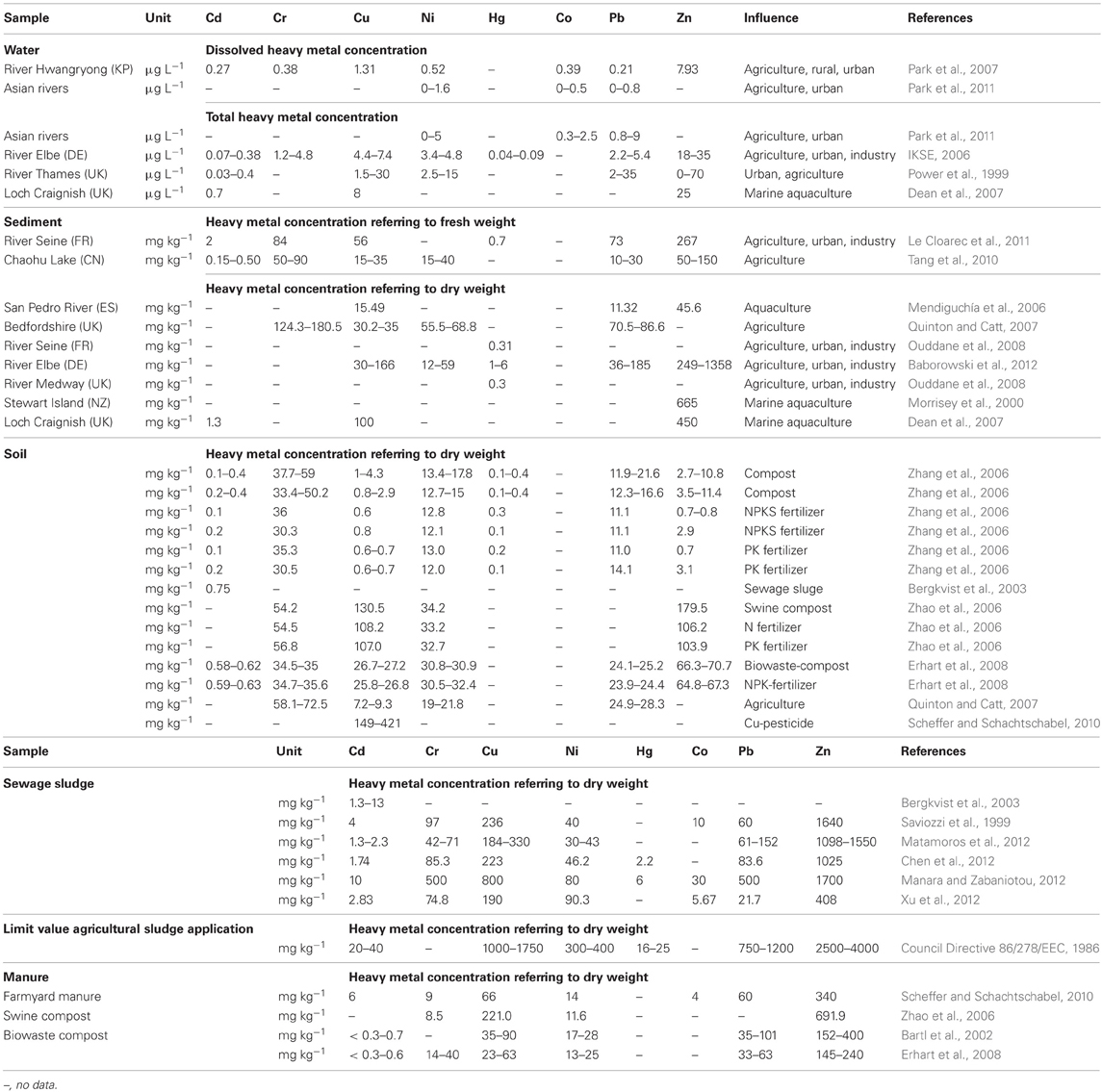
Fish farmers frequently use pharmaceuticals (such as antibiotics) and metal containing products to prevent fouling, to feed and to treat fish in order to limit the spread of infections (Burridge et al., 2010). For instance, copper (Cu)-containing materials are applied as anti-fouling agents for farm cages and nets; some cages themselves are made from Cu alloys (Burridge et al., 2010). Therefore, bacterial communities of aquacultures are strongly exposed to the combination of heavy metals and antibiotics. The exposure to both antimicrobial substances may increase the likelihood of selection and co-selection of antibiotic resistance. Moreover, the high nutritional value and the relatively low cost of wastewater, excreta, and sewage sludge convert such heavy metal containing waste to valuable fish feed, especially in developing countries (WHO, 2006). The relevance of heavy metal contaminations in aquaculture has been illustrated by Choi and Cech (1998), who found unexpected high concentrations of mercury (Hg) in fish feed. The enrichment of aquaculture sediments with zinc (Zn) (Morrisey et al., 2000) and Cu (Smith et al., 2005; Burridge et al., 2010) as well as cadmium (Cd) (Dean et al., 2007) and lead (Pb) (Mendiguchía et al., 2006) was reported earlier and is presented in Table 1.
Most serious heavy metal contaminations in soils include Cu, Hg, Zn, Pb, and Cd (Han et al., 2002). Land application of metal containing fertilizers, sewage sludge, and liquid manure is common practice in agriculture not only in Europe but also in other regions of the world. Due to those applications heavy metals such as Pb, Hg, Cd, Cu, Zn, chromium (Cr), and nickel (Ni) are transferred to arable soil (Table 1). Because of its bactericidal and fungicidal properties, Cu-containing pesticides are applied in organic and conventional agriculture (Nemecek et al., 2011). Furthermore, metals such as iron (Fe), cobalt (Co), manganese (Mn), Cu, and Zn are applied as nutritional additives in animal feed for livestock farming and fish production in Europe (Commission Regulation 1831/2003/EC, 2003).
A pre-assessment of the environmental impact of Zn and Cu feed amendments in the European Union demonstrated the major role of aquaculture and agriculture as pollution sources of those metals. The applied models predicted that the no effect concentrations of Cu and Zn will be exceeded in some soil and water systems within the next 10–50 years (Monteiro et al., 2010). Moreover, agriculture was identified as the main source of Cu- and Zn-contamination of arable soil in England and Wales (Nicholson et al., 2003). In addition, 30% of the Cd input to the investigated agricultural soil originated from inorganic fertilizers.
Heavy Metal Toxicity and Resistance
Not all heavy metals are equally toxic to bacteria. Some are important trace metals involved in various physiological functions of the cell. For example Zn, Ni, Cr, Cu, and Co are metals of moderate to high physiological importance. They are essential micronutrients necessary for several cellular functions and components of DNA- and RNA-polymerases (Zn), urease (Ni), cytochrome (Cr) and cytochrom—c—oxidase (Cu). Pb, Cd, Hg, silver (Ag), and gold (Au) have reduced relevance as trace nutrients and they have limited physiological function. Cd and Hg are strong cellular toxins because of their ability to form harmful complexes (Nies, 1999). In contrast, the toxicity of trace metals such as Zn, Ni, Cu, Co, and Cr are strongly dependent on the concentration. As reviewed by Nies (1999), the elements Fe, Mn and molybdenum (Mo) were described as physiologically important with limited toxicity. Metals such as Zn, Ni, Cu, Co, Cr, vanadium (V), and tungsten (W) are toxic elements with metabolic relevance, while the elements Ag, Cd, Hg, Pb, antimony (Sb), and uranium (U) are strong toxins.
The toxicity of heavy metals in the environment strongly depends on the environmental conditions because these conditions influence the valence of the metal ions and therefore their bioavailability. Environmental Cr, for example mainly occurs in two different forms: as Cr3+ ion or as the hexavalent Cr associated with oxygen as chromate (for example CrO2−4). The Cr3+ ions are less toxic to bacteria than the chromate (Nies, 1999). Environmental conditions like the pH-value, the concentration of organic matter and the redox potential affect the concentrations and bioavailability of heavy metals in soil, sediment, and water. For instance, the oxygen level influences the redox potential and thereby affects the solubility of some metals. In some water bodies the decomposition of high concentrations of organic matter leads to a reduction of the oxygen level down to anaerobic conditions. Under such conditions the solubility of Cd and Zn is reduced (Schulz-Zunkel and Krueger, 2009). On the other hand, low pH-values increase the solubility of the metals Pb, Cd, and Zn. High contents of organic matter within the sediment act as a sink for some metals: for example Cr and Zn are known to bind to organic matter (Schulz-Zunkel and Krueger, 2009).
In general, the microbial toxicity of heavy metals is due to their chemical affinity to the thiol groups and macrobiomolecules but also depends on the solubility of the metal compound under physiological conditions (Nies, 1999). To avoid cellular damage caused by metal ions, bacteria evolved mechanisms of metal tolerance. There are three general mechanisms which result in heavy metal resistance: the first mechanism is the complex formation or sequestration of toxic metals (Silver and Phung, 1996). Upon metal binding, the concentration of the free toxic ions in the cytoplasm is minimized. Biosorption of toxic metals is known from cell membranes, cell walls and extracellular polymeric substance (EPS) of biofilms (Harrison et al., 2007). For example, the EPS matrix and the contained polysaccharides were reported to bind heavy metals (Teitzel and Parsek, 2003). Thus, the metal tolerance of the bacteria belonging to that biofilm was enhanced. The second mechanism of resistance to toxic metals is the detoxification through reduction of intracellular ions (Nies, 1999). A well understood example is the mercury reductase, encoded by the merA gene. This MerA protein reduces Hg2+ to the less toxic Hg0 (Schiering et al., 1991). Hg0 will then diffuse out of the cell, due to its low evaporation point (Nies, 1999). Finally, extrusion of toxic ions by efflux systems is the third mechanism of heavy metal resistance (Nies and Silver, 1995). The cation/proton antiporter Czc, known for example from Alcaligenes eutrophus, mediates resistance to the metal ions Cd2+, Zn2+, and Co2+ by extrusion of metals from the cytoplasm though the inner and outer membrane to the surrounding environment (Silver and Phung, 1996). Population wide metal tolerance is increased by persister cells (Harrison et al., 2007). Persister cells mediate time dependent tolerance to toxic metal ions due to upregulation of resistance and stress response genes (Harrison et al., 2007).
Bacterial sensitivity can be quite complex, nevertheless, some generalizations seem to be possible. Gram positive bacteria are described to be more sensitive to toxic metals than gram negative bacteria (Sterritt and Lester, 1980). Moreover, two general microbial toxicity rankings were reported (Nies, 1999; Harrison et al., 2007). In these rankings bacterial susceptibility is described as a function of the particular metal sulfide dissociation constants (pKSP) (Nies, 1999) and as a function of the standard reduction potentials (ΔE0) (Harrison et al., 2007). Those two general toxicity rankings are shown in Table 2. Nevertheless, different types of bacteria show different sensitivities to toxic metals. Even the heavy metal susceptibility of bacteria belonging to the same genera can differ dramatically. As an example, while the growth of one Aeromonas isolate is inhibited by a concentration of 100 μg Zn ml−1, another strain of the same genera isolated from the same sampling site has the ability to grow up to a concentration of 3200 μg Zn ml−1 (Matyar et al., 2010). Further examples showing bacteria and their susceptible to toxic metals are displayed in Table 2. The listed examples did not show the same pattern of toxicity as reported by Nies (1999) and Harrison et al. (2007). This demonstrates that environmental bacteria may adapt to their ecological conditions and may have been selected for certain metal tolerance mechanisms.
The Machinery of Co-Selection
Since the 1970's, there has been great concern about heavy metals selecting indirectly for antibiotic resistance by co-selection (Koditschek and Guyre, 1974). This indirect selection process is due to a coupling of the resistance mechanisms against antibiotics and heavy metals. Those mechanisms can be coupled physiologically (cross-resistance) and genetically (co-resistance). Cross-resistance describes mechanisms that provide tolerance to more than one antimicrobial agent such as antibiotics and heavy metals (Chapman, 2003). As an example, several multi drug efflux pumps are known to mediate decreased susceptibility toward antibiotics and heavy metals by rapid extrusion of the toxins out of the cell (Martinez et al., 2009). Further well-characterized cross-resistance mechanisms were reviewed by Baker-Austin et al. (2006). Co-resistance is defined as two or more genetically linked resistance genes, meaning that genes responsible for two or more resistances are located next to each other on one mobile genetic element (Chapman, 2003). As an example, Osman et al. (2010) isolated an aquatic bacterium harbouring a plasmid which contained genes conferring resistance to antibiotics and metals like Cr and Co. Due to the close arrangement of the genes it is likely that these genes are subject to a combined transmission in the case of a horizontal gene transfer. A genetic linkage of Cu resistance encoded by the tcrB gene, macrolide [erm(B)] and glycopeptid resistance (vanA) was observed in Enterococcus faecium isolated from farm animals (Hasman and Aarestrup, 2002). Here co-resistance to Cu and antibiotics, all applied in farming practice was detected. Macrolides are commonly used in veterinary medicine (Grave et al., 2010) and glycopeptide antibiotics have been used as growth promoters for animal production in the past. Nowadays glycopeptide antibiotics, such as vancomycin, belong to the group of last resort antibiotics in human medicine. Thus, this genetic linkage found by Hasman and Aarestrup (2002) could be an example for a Cu-induced spread of resistance to antibiotics relevant in veterinary and human medicine. Furthermore, Aeromonas salmonicida subsp. salmonicida isolated from Atlantic salmon (Salmo salar) from aquaculture facilities was identified carrying Hg (mer operon) and multiple antibiotic resistance genes (aadA7, sulI, sulII, floR, tetA, tetR, strA, strB, and blaCMY−2) on an IncA/C plasmid (McIntosh et al., 2008). This was the first finding of plasmid associated resistance to florfenicol (floR), an antibiotic usually used to treat furunculosis in aquacultures.
Integrons are genetic elements capable of acquiring and exchanging DNA fragments named gene cassettes. Furthermore, class 1 integrons are assumed to catalyse co-selection because they frequently contain gene cassettes that mediate resistance to antibiotics and they are frequently found in contaminated habitats. Presumably these integrons mediate a selective advantage to bacteria that occur under stressful environmental conditions, for example due to toxic metals. This assumption is supported by several studies which discovered elevated abundance of class 1 integrons in aquatic environments contaminated with heavy metals (Wright et al., 2008; Rosewarne et al., 2010). Further indications for a co-resistance mechanism in fresh water bacteria were given by Gillings et al. (2008) and Stokes et al. (2006). Both publications document class 1 integrons which are closely located to genes coding for the multi drug efflux pump czcA. This efflux pump is known to extrude the metal ions Zn2+, Cd2+, and Co2+.
Moreover, environmental pollution by heavy metals not only triggers co-selection processes, but also increases the level of tolerance to antibiotics due to co-regulation of resistance genes. Heavy metal ions are known to co-regulate genes responsible for antibiotic resistance and decrease antibiotic susceptibility (Baker-Austin et al., 2006). For example, the soxS protein is a regulator for the AcrAB efflux system in Escherichia coli. Under oxidative stress, for instance caused by several metal ions like Cr2O−7 and Cu2+, soxS is upregulated (Harrison et al., 2009). The subsequently increased production of the AcrAB efflux system additionally mediates enhanced tolerance toward antibiotics such as chloramphenicol, tetracyclin, novobiocin, nafcillin, and oxacillin.
Studies investigating co-selection in the environment frequently show the correlation of increased heavy metal concentrations with increased phenotypic or genotypic antibiotic resistance (Table 3). However, some studies indicate that increasing heavy metal concentrations lead to a decrease of antibiotic resistance (Stepanauskas et al., 2005; Tuckfield and McArthur, 2008; Hölzel et al., 2012). These contradicting results were investigated by Hölzel et al. (2012). In consequence of the addition of mercury chloride (HgCl2) to the antimicrobial test procedure the minimum inhibitory concentration (MIC) for a wide range of antibiotics decreased. This observation could be due to an interaction of Hg with enzymes or nucleic acids which cause antibiotic resistance. HgCl2 could also have a co-toxic effect with antibiotics that interfere with ribosomes because the regeneration of the Hg-degraded enzyme would be inhibited. Furthermore, Hölzel et al. (2012) mentioned also a possible metal induced shift within the bacterial community toward Hg tolerant bacteria whereby the benefit of antibiotic resistance in the presence of antibiotics would be outcompeted. The increased antibiotic susceptibility in consequence of Hg exposure could also play a role in the observations of other field studies. For example, Tuckfield and McArthur (2008) observed decreasing microbial aminoglycoside resistance at sites with increased Hg concentrations.
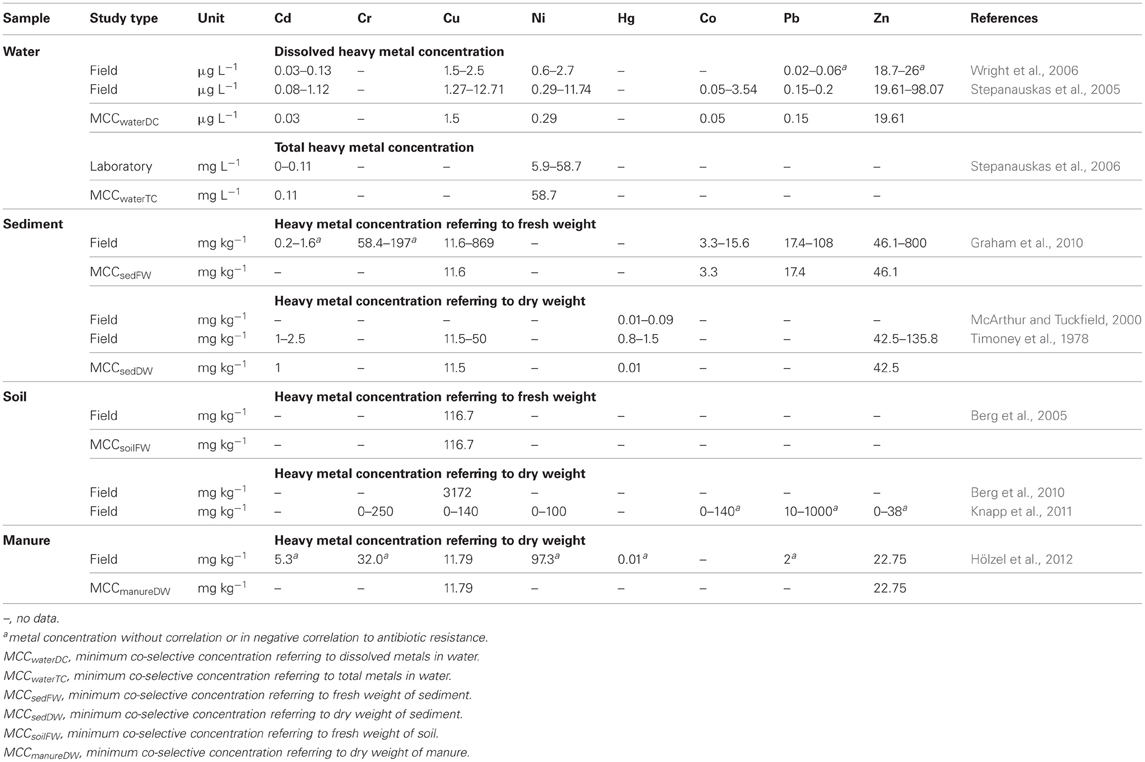
Table 3. Heavy metal concentrations of studies investigating co-selection in laboratory and field experiments.
Risk Assessment for Metal Driven Co-Selection
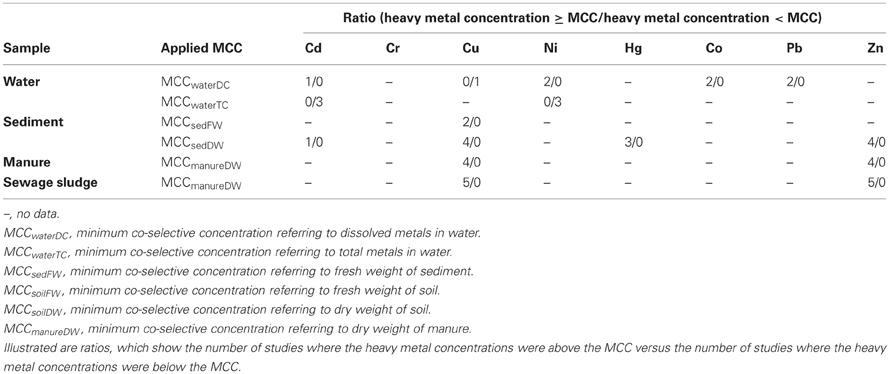
To assess the risk for the co-selection of antibiotic and heavy metal resistance, two datasets of heavy metal concentrations were compared. One of the datasets is shown in Table 3, containing metal concentrations observed in studies that investigated co-selection in laboratory and field experiments. The second dataset is shown in Table 1, containing heavy metal concentrations that were measured in various environmental compartments which are impacted by agriculture and/or aquaculture. Additionally, we adapted a concept of the MIC originating from the antimicrobial susceptibility testing in clinical settings. The MIC is defined as the antibiotic concentration that is needed to inhibit bacterial growth. If this MIC has been increased above an epidemiological cut-off value of a bacterial strain the strain will be defined as antibiotic (microbiological) resistant (URL: http://www.eucast.org). In this context the minimum heavy metal concentration which correlates with a detection of increased bacterial antibiotic resistance, was specified as the minimum co-selective concentration (MCC) of a metal (Table 3). Additionally, specific MCCs to every environmental compartment and the respective analytic detection method of the metals (for example for dissolved and total metal concentrations or metal content referring to dry or fresh weight of the solid samples) were defined (Table 3). Moreover, the MCCs for each metal were subsequently compared to the heavy metal levels found within the corresponding environmental compartments (Table 4). Environmental metal concentrations that exceeded the corresponding MCC were considered as potential drivers of co-selection of antibiotic resistance in the environment.
In the natural water environment (water and sediment) Cd, Cu, Ni, Hg, Co, Pb, and Zn frequently reach levels that exceed their respective MCC values (Table 4) and therefore, may drive co-selection. While there are several studies available investigating co-selection in the water environment, there are only a few publications considering soil environments (Berg et al., 2005, 2010; Knapp et al., 2011). Thus, the assessment of the risk for the co-selection of antibiotic and heavy metal resistance in soil is limited to Cr, Cu, and Ni (Table 3). Furthermore, the data of Knapp et al. (2011) does not allow extracting MCCs of metals because the lowest metal concentrations that may caused the increase in antibiotic resistance gene abundance are not shown. However, these results provide evidence for Cr, Cu, and Ni driven co-selection of antibiotic resistance in soil. Some Cu and Cr levels of the reviewed arable soil samples (Table 1) are similar or even higher than the levels observed by Knapp et al. (2011) (Table 3). Sewage sludge and manure are part of this risk assessment because those organic fertilizers themselves could facilitate metal driven co-selection of antibiotic resistance before entering soil environments and they might additionally transfer metals to arable soil. Cu and Zn concentration of sewage sludge and manure frequently exceeded the MCCs of manure (Table 4). Moreover, the limit values for heavy metal concentrations of sewage sludge for the use in agriculture (Council Directive 86/278/EEC, 1986) are much higher than the MCCs of manure (Tables 1 and 3). As mentioned earlier in this article, the use of Zn and Cu in animal farming and agriculture is common and those metals have been investigated in all considered environmental compartments. The concentrations for both metals exceed their MCCs for some water, sediment, sewage sludge, and manure samples. In soil Cu levels reach concentrations that are reported as potentially co-selective for antibiotic resistance genes (Knapp et al., 2011). In contrast, a Zn MCC for soil samples could not be evaluated because Knapp et al. (2011) did not detect increasing abundance of antibiotic resistance genes in correlation with elevated Zn concentrations. However, the Zn concentrations of soil samples investigated by Knapp et al. (2011) were relatively low compared to other soils (Table 1) and maybe within the no effect range. In summary, all considered heavy metals (frequently Cu and Zn) reach concentrations above their MCCs in the different environmental compartments. Therefore, the analysis of the data suggests that heavy metal concentrations in soil and water bodies occasionally reach levels that might drive a co-selection of antibiotic resistance.
This risk assessment of heavy metal driven co-selection is based on MCCs which are derived from positive correlations of increased metal concentrations with increased antibiotic resistance. This risk assessment provides a tool to estimate at which levels environmental metal concentrations may cause the dissemination of microbial antibiotic resistance due to co-selection. Ideally such a risk assessment would be conducted under laboratory conditions, as it is currently the case for the determination of the MIC. For the purpose of this study this was not possible as we wanted to review existing studies and included laboratory and field data, in order to detect a first pattern or synthesis on heavy metal induced co-selection of antibiotic resistance in the field. The results of the MCC analysis for such data need to be carefully interpreted, mainly because positive correlations between metal levels and antibiotic resistance could also be false positive. This would be the case, if another selection pressure (for example by antibiotics) would be the trigger of the observed selection and not the co-occurring metal.
In order to better assess the risk of co-selection of antibiotic and heavy metal resistance more research is necessary. Only a limited number of studies is available that investigated the co-selection in water and soil environments and additionally measured heavy metal concentrations (Table 3). Especially for soil environments, there is only one multi metal study on co-selection and metal contamination (Knapp et al., 2011). The knowledge about the natural background of antibiotic resistance gene abundance (resistome) in the different environments is also limited. Thus, we cannot distinguish between the natural resistome and an elevated abundance of antibiotic resistance genes in different environmental samples. Therefore, it is difficult to detect an increase of antibiotic resistance genes in field studies. Further research should relate to the MCCs of heavy metals. Although we were able to derive the MCC values form recent studies, further research in field and laboratory experiments is urgently requested to broaden the database of co-selective concentrations. Stepanauskas et al. (2006) investigated such a selective concentration of Ni and Cd in a lab experiment. Nevertheless, the metal concentrations observed as minimum selective concentrations in those microcosms were much higher compared to the Ni and Cd levels observed in environmental water samples (Tables 1 and 3). This difference requires further investigation and may be an artifact of the growth in the laboratory since laboratory grown bacteria usually have better conditions than their environmental counterparts.
While our MCC approach provides a first step toward a unifying concept for analyzing co-selection of antibiotic resistance through heavy metals, there is an urgent need to extend this approach to a comprehensive risk assessment. The need of such a risk assessment is illustrated in our results, which show that in all considered environmental compartments (water, sediment, and soil) as well as sewage sludge and manure, one or more heavy metals reach concentrations that may lead to a metal driven co-selection of antibiotic resistance.
Conclusion
Concluding all these facts concerning the heavy metal driven co-selection of antibiotic resistance, metals such as Cd, Hg, Cu, and Zn are of great importance in water and soil environments that are influenced by agriculture and aquaculture. These metals are moderately to highly toxic to bacteria; they reach the environment and they accumulate to selective concentrations. Additionally, they can trigger co-selection of antibiotic resistance because responsible co-selection mechanisms that mediate resistance to these heavy metals and clinically as well as veterinary relevant antibiotics have already been described. Therefore, the elimination of antibiotics from the list of animal feed additives as growth promoters was a step in the right direction. Further steps need to be taken to reduce the alarming spread of antibiotic resistance genes. In addition to the avoidance of antibiotics in livestock farming and aquaculture, further antimicrobial agents such as heavy metals should be considered. These metals have the potential to act as a selective pressure that forces the proliferation and evolution of antibiotic and heavy metal resistance in the natural environment. With the exception of the above mentioned studies, investigations which explicitly test for the co-selection of heavy metals and antibiotics used in animal farms and aquaculture are still scarce. Future studies investigating heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture should focus on Hg, Cd, Cu, and Zn as co-selecting factors for the evolution of antibiotic resistances. Nevertheless, the respective environmental background has to be taken into account.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was kindly supported by Helmholtz Impulse and Networking Fund through Helmholtz Interdisciplinary Graduate School for Environmental Research (HIGRADE) and the funding of the BMBF programs “IWAS” and “ANTIRESIST”. The authors also thank Anka Broßio and Shane Denecke for linguistic support.
References
Abskharon, R. N. N., Hassan, S. H. A., Gad El-Rab, S. M. F., and Shoreit, A. A. M. (2008). Heavy metal resistant of E. coli isolated from wastewater sites in Assiut City, Egypt. Bull. Environ. Contam. Toxicol. 81, 309–315.
Akinbowale, O. L., Peng, H., Grant, P., and Barton, M. D. (2007). Antibiotic and heavy metal resistance in motile Aeromonads and Pseudomonads from Rainbow trout (Oncorhynchus mykiss) farms in Australia. Int. J. Antimicrob. Agents 30, 177–182.
Alanis, A. J. (2005). Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36, 697–705.
Aminov, R. I. (2010). A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbio. 1:134. doi: 10.3389/fmicb.2010.00134
Baborowski, M., Büttner, O., Morgenstern, P., Jancke, T., and Westrich, B. (2012). Spatial variability of metal pollution in groyne fields of the Middle Elbe—implications for sediment monitoring. Environ. Pollut. 167, 115–123}.
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182.
Bartl, B., Hartl, W., and Horak, O. (2002). Long-term application of biowaste compost versus mineral fertilization: effects on the nutrient and heavy metal contents of soil and plants. J. Plant Nutr. Soil Sci. 165, 161–165.
Berg, J., Thorsen, M. K., Holm, P. E., Jensen, J., Nybroe, O., and Brandt, K. K. (2010). Cu exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 44, 8724–8728.
Berg, J., Tom-Petersen, A., and Nybroe, O. (2005). Copper amendment of agricultural soil selects for bacterial antibiotic resistance in the field. Lett. Appl. Microbiol. 40, 146–151.
Bergkvist, P., Jarvis, N., Berggren, D., and Carlgren, K. (2003). Long-term effects of sewage sludge applications on soil properties, cadmium availability and distribution in arable soil. Agric. Ecosyst. Environ. 97, 167–179.
Boxall, A. B. A., Blackwell, P., Cavallo, R., Kay, P., and Tolls, J. (2002). The sorption and transport of a sulphonamide antibiotic in soil systems. Toxicol. Lett. 131, 19–28.
Burridge, L., Weis, J. S., Cabello, F., Pizarro, J., and Bostick, K. (2010). Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture 306, 7–23.
Chapman, J. S. (2003). Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegradation 51, 271–276.
Chen, H., Yan, S. H., Ye, Z. L., Meng, H. J., and Zhu, Y. G. (2012). Utilization of urban sewage sludge: Chinese perspectives. Environ. Sci. Pollut. Res. 19, 1454–1463.
Choi, M. H., and Cech, J. J. (1998). Unexpectedly high mercury level in pelleted commercial fish feed. Environ. Toxicol. Chem. 17, 1979–1981.
Commission Regulation 1831/2003/EC. (2003). Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003 on additives for use in animal nutrition. Off. J. Eur. Union L268, 29–43.
Council Directive 86/278/EEC. (1986). Council Directive 86/278/EEC of 12 June 1986 on the protection of the environment, and in particular of the soil, when sewage sludge is used in agriculture. Off. J. Eur. Comm. L181, 6–12.
Dean, R. J., Shimmield, T. M., and Black, K. D. (2007). Copper, zinc and cadmium in marine cage fish farm sediments: an extensive survey. Environ. Pollut. 145, 84–95.
Erhart, E., Hartl, W., and Putz, B. (2008). Total soil heavy-metal concentrations and mobile fractions after 10 years of biowaste-compost fertilization. J. Plant Nutr. Soil Sci. 171, 378–383.
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111.
Gillings, M., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., et al. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190, 5095–5100.
Graham, D., Olivares-Rieumont, S., Knapp, C., Lima, L., Werner, D., and Bowen, E. (2010). Antibiotic resistance gene abundances associated with waste discharges to the Almendares river near Havana, Cuba. Environ. Sci. Technol. 45, 418–424.
Grave, K., Torren-Edo, J., and Mackay, D. (2010). Comparison of the sales of veterinary antibacterial agents between 10 European countries. J. Antimicrob. Chemother. 65, 2037–2040.
Han, F., Kingery, W., and Selim, H. (2001). “Accumulation, redistribution, transport and bioavailability of heavy metals in wasteamended soils,” in Trace Elements in Soil: Bioavailability, Fluxes and Transfer, eds I. Iskander and M. Kirkham (Boca Raton, FL: CRC), 141–168.
Han, F. X., Banin, A., Su, Y., Monts, D. L., Plodinec, J. M., Kingery, W. L., et al. (2002). Industrial age anthropogenic inputs of heavy metals into the pedosphere. Naturwissenschaften 89, 497–504.
Harrison, J. J., Ceri, H., and Turner, R. J. (2007). Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5, 928–938.
Harrison, J. J., Tremaroli, V., Stan, M. A., Chan, C. S., Vacchi-Suzzi, C., Heyne, B. J., et al. (2009). Chromosomal antioxidant genes have metal ion-specific roles as determinants of bacterial metal tolerance. Environ. Microbiol. 11, 2491–2509.
Hasman, H., and Aarestrup, F. M. (2002). tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46, 1410–1416.
Heuer, H., Schmitt, H., and Smalla, K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243.
Hölzel, C. S., Müller, C., Harms, K. S., Mikolajewski, S., Schäfer, S., Schwaiger, K., et al. (2012). Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ. Res. 113, 21–27.
Knapp, C. W., McCluskey, S. M., Singh, B. K., Campbell, C. D., Hudson, G., and Graham, D. W. (2011). Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived scottish soils. PLoS ONE 6:e27300. doi: 10.1371/journal.pone.0027300
Koditschek, L. K., and Guyre, P. (1974). Resistance transfer fecal coliforms isolated from the Whippany River. Water Res. 8, 747–752.
Le Cloarec, M. F., Bonte, P. H., Lestel, L., Lefèvre, I., and Ayrault, S. (2011). Sedimentary record of metal contamination in the Seine River during the last century. Phys. Chem. Earth Parts A/B/C 36, 515–529.
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129.
Malik, A., and Aleem, A. (2011). Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environ. Monit. Assess. 178, 293–308.
Manara, P., and Zabaniotou, A. (2012). Towards sewage sludge based biofuels via thermochemical conversion –a review. Renew. Sust. Energ. Rev. 16, 2566–2582.
Martinez, J. L., Sánchez, M. B., Martínez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449.
Matamoros, V., Nguyen, L. X., Arias, C. A., Nielsen, S., Laugen, M. M., and Brix, H. (2012). Musk fragrances, DEHP and heavy metals in a 20 years old sludge treatment reed bed system. Water Res. 46, 3889–3896.
Matyar, F., Akkan, T., Uçak, Y., and Eraslan, B. (2010). Aeromonas and Pseudomonas: antibiotic and heavy metal resistance species from Iskenderun Bay, Turkey (northeast Mediterranean Sea). Environ. Monit. Assess. 167, 309–320.
McArthur, J. V., and Tuckfield, R. C. (2000). Spatial patterns in antibiotic resistance among stream bacteria: effects of industrial pollution. Appl. Environ. Microbiol. 66, 3722–3726.
McIntosh, D., Cunningham, M., Ji, B., Fekete, F. A., Parry, E. M., Clark, S. E., et al. (2008). Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 61, 1221–1228.
Mendiguchía, C., Moreno, C., Mánuel-Vez, M. P., and García-Vargas, M. (2006). Preliminary investigation on the enrichment of heavy metals in marine sediments originated from intensive aquaculture effluents. Aquaculture 254, 317–325.
Monteiro, S. C., Lofts, S., and Boxall, A. B. A. (2010). Pre-Assessment of Environmental Impact of Zinc and Copper Used in Animal Nutrition. Parma, Italy: European Food Safety Authority.
Morrisey, D. J., Gibbs, M. M., Pickmere, S. E., and Cole, R. G. (2000). Predicting impacts and recovery of marine-farm sites in Stewart Island, New Zealand, from the Findlay–Watling model. Aquaculture 185, 257–271.
Nemecek, T., Dubois, D., Huguenin-Elie, O., and Gaillard, G. (2011). Life cycle assessment of Swiss farming systems: I. Integr. Org. Farm. Agric. Syst. 104, 217–232.
Nicholson, F. A., Smith, S. R., Alloway, B. J., Carlton-Smith, C., and Chambers, B. J. (2003). An inventory of heavy metals inputs to agricultural soils in England and Wales. Sci. Total Environ. 311, 205–219.
Nies, D. H., and Silver, S. (1995). Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14, 186–199.
Osman, O., Tanguichi, H., Ikeda, K., Park, P., Tanabe-Hosoi, S., and Nagata, S. (2010). Copper-resistant halophilic bacterium isolated from the polluted Maruit Lake, Egypt. J. Appl. Microbiol. 108, 1459–1470.
Ouddane, B., Mikac, N., Cundy, A. B., Quillet, L., and Fischer, J. C. (2008). A comparative study of mercury distribution and methylation in mudflats from two macrotidal estuaries: the Seine (France) and the Medway (United Kingdom). Appl. Geochem. 23, 618–631.
Park, J.-H., Inam, E., Abdullah, M. H., Agustiyani, D., Duan, L., Hoang, T. T., et al. (2011). Implications of rainfall variability for seasonality and climate-induced risks concerning surface water quality in East Asia. J. Hydrol. 400, 323–332.
Park, J.-H., Lee, J.-H., Kang, S.-Y., and Kim, S.-Y. (2007). Hydroclimatic controls on dissolved organic matter (DOM) characteristics and implications for trace metal transport in Hwangryong River Watershed, Korea, during a summer monsoon period. Hydrol. Process. 21, 3025–3034.
Poirel, L., Kampfer, P., and Nordmann, P. (2002). Chromosome-encoded ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040.
Poirel, L., Rodriguez-Martinez, J.-M., Mammeri, H., Liard, A., and Nordmann, P. (2005). Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49, 3523–3525.
Power, M., Attrill, M. J., and Thomas, R. M. (1999). Heavy metal concentration trends in the Thames estuary. Water Res. 33, 1672–1680.
Quinton, J. N., and Catt, J. A. (2007). Enrichment of heavy metals in sediment resulting from soil erosion on agricultural fields. Environ. Sci. Technol. 41, 3495–3500.
Rosewarne, C. P., Pettigrove, V., Stokes, H. W., and Parsons, Y. M. (2010). Class 1 integrons in benthic bacterial communities: abundance, association with Tn402-like transposition modules and evidence for coselection with heavy-metal resistance. FEMS Microbiol. Ecol. 72, 35–46.
Salyers, A. A., and Amábile-Cuevas, C. F. (1997). Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41, 2321–2325.
Saviozzi, A., Biasci, A., Riffaldi, R., and Levi-Minzi, R. (1999). Long-term effects of farmyard manure and sewage sludge on some soil biochemical characteristics. Biol. Fertil. Soils 30, 100–106.
Scheffer, F., and Schachtschabel, P. (2010). “Gefährdung der Bodenfunktionen,” in Lehrbuch der Bodenkunde, eds H. Lieth and B. Markert (Heidelberg: Spektrum Akademischer Verlag), 449–519.
Schiering, N., Kabsch, W., Moore, M. J., Distefano, M. D., Walsh, C. T., and Pai, E. F. (1991). Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain-RC607. Nature 352, 168–172.
Schulz-Zunkel, C., and Krueger, F. (2009). Trace metal dynamics in floodplain soils of the River Elbe: a review. J. Environ. Qual. 38, 1349–1362.
Silver, S., and Phung, L. T. (1996). Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50, 753–789.
Smith, J. N., Yeats, P. A., and Milligan, T. G. (2005). “Sediment geochronologies for fish farm contaminants in Lime Kiln Bay, Bay of Fundy,” in Handbook of Environmental Chemistry, ed B. T. Hargrave (Berlin: Springer), 221–238.
Sørum, H. (2006). “Antimicrobial drug resistance in fish pathogens,” in Antimicrobial Resistance in Bacteria of Animal Origin, ed F. M. Aarestrup (Washington, DC: ASM Press), 213–238.
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., King, C. J., et al. (2006). Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8, 1510–1514.
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., and McArthur, J. V. (2005). Elevated microbial tolerance to metals and antibiotics in metal-contaminated industrial environments. Environ. Sci. Technol. 39, 3671–3678.
Sterritt, R. M., and Lester, J. N. (1980). Interactions of heavy metals with bacteria. Sci. Total Environ. 14, 5–17.
Stokes, H. W., Nesbo, C. L., Holley, M., Bahl, M. I., Gillings, M. R., and Boucher, Y. (2006). Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 188, 5722–5730.
Tang, W., Shan, B., Zhang, H., and Mao, Z. (2010). Heavy metal sources and associated risk in response to agricultural intensification in the estuarine sediments of Chaohu Lake Valley, East China. J. Hazard. Mater. 176, 945–951.
Teitzel, G. M., and Parsek, M. R. (2003). Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69, 2313–2320.
Timoney, J. F., Port, J., Giles, J., and Spanier, J. (1978). Heavy-metal and antibiotic resistance in the bacterial flora of sediments of New York bight. Appl. Environ. Microbiol. 36, 465–472.
Tuckfield, R. C., and McArthur, J. V. (2008). Spatial analysis of antibiotic resistance along metal contaminated streams. Microb. Ecol. 55, 595–607.
WHO. (2006). Guidelines for the Safe Use of Wastwater, Excreta and Greywater. Volume 3: Wastewater and Excreta Use in Aquaculture. Geneva: World Health Organization.
Wright, M. S., Baker-Austin, C., Lindell, A. H., Stepanauskas, R., Stokes, H. W., and McArthur, J. V. (2008). Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2, 417–428.
Wright, M. S., Peltier, G. L., Stepanauskas, R., and McArthur, J. V. (2006). Bacterial tolerances to metals and antibiotics in metal-contaminated and reference streams. FEMS Microbiol. Ecol. 58, 293–302.
Xu, J.-Q., Yu, R.-L., Dong, X.-Y., Hu, G.-R., Shang, X.-S., Wang, Q., et al. (2012). Effects of municipal sewage sludge stabilized by fly ash on the growth of Manilagrass and transfer of heavy metals. J. Hazard. Mater. 217–218, 58–66.
Zhang, M., Heaney, D., Henriquez, B., Solberg, E., and Bittner, E. (2006). A four-year study on influence of biosolids/MSW cocomposta application in less productive soils in Alberta: nutrient dynamics. Compost Sci. Util. 14, 68–80.
Keywords: co-selection, antibiotic resistance, heavy metal, agriculture, aquaculture, farming
Citation: Seiler C and Berendonk TU (2012) Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbio. 3:399. doi: 10.3389/fmicb.2012.00399
Received: 01 March 2012; Accepted: 30 October 2012;
Published online: 14 December 2012.
Edited by:
Rustam I. Aminov, University of the West Indies, JamaicaReviewed by:
Henning Sørum, Norwegian School of Veterinary Science, NorwayJan Siemens, University of Bonn, Germany
Copyright © 2012 Seiler and Berendonk. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Claudia Seiler, Institute of Hydrobiology, Technische Universität Dresden, Zellescher Weg 40, 01217 Dresden, Germany. e-mail: claudia.seiler@mailbox.tu-dresden.de